Research areas in the Cheng Zhang’s Group mainly focus on the complex DNA molecular circuit and computing, DNA assembly assisted single molecular nanopore detection, programmable DNA/nanoparticle assembly, single molecular FRET, super-resolution single molecular analysis, DNA origami nanodevice design and fabrication, and applications development for DNA molecular control and engineering.
Our research is multidisciplinary, involving concepts and expertise from bioengineering, chemistry, material science, physics, and electrical engineering, mathematics, computer science. The synthesis and design of DNA assembled nanostructures and nanodevices is carried out in multiple steps including wet experiments and model simulations, where computer assisted design and analysis are often required. Using these well designed DNA nanostructures and circuits, we further want to explore them to the intracellular environments. The nanodevices of current interest are iterative complex molecular circuit and nanodevice, programmable assembled nanopore analysis, addressable nanoparticles assembly, nanoparticle based intracellular detection and origami based super-resolution image.
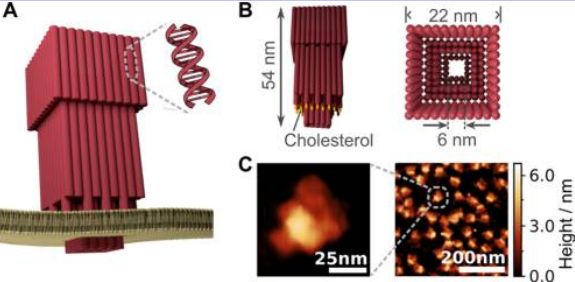
DNA assembly based tunable nanopore analysis
We are developing new nanopore technologies combined with DNA assembly and nanopore to sensitively analysis of the single molecule translocations. We particularly interested in the assembled origami carrier escorting nanopore translocation and synthetic assembled nanopore. Via DNA origami carrier, more diverse spatial cargo loading modes can be achieved. In addition, assembled origami nanopore can facilitate translocation of various molecules with different sizes and shapes. We envision that DNA assembly based nanopore method will find more applications in the areas of nanopore based molecular sensing and detection.
DNA circuit: operation, computing and analysis
We are now interested in developing complex molecular circuits and computing operations to facilitate signal transmission and the applications in vitro and in cellular environment. Currently, we are working on: recyclable complex DNA circuit, DNAzyme regulated molecular circuits, cascading transcription circuits. Currently we are working on integrating the in vitro DNA circuit into cells, thus achieving in vivo molecular diagnosis and information signaling in cell. In addition, regulable DNA circuit can also be used to construct dynamic nanodevices, which is another interest in our research.

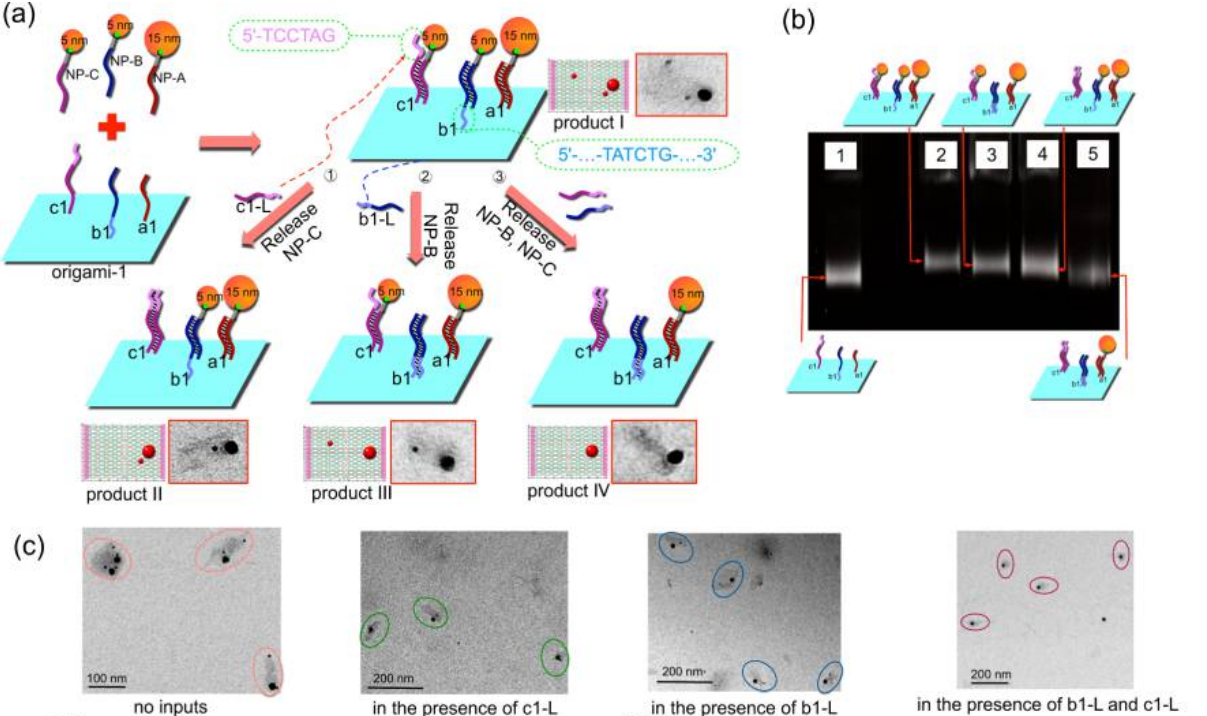
DNA directed nanoparticle assembly, arrangement and modification
Because of the unique structural and optical properties, organizing AuNPs with DNA assembly structures in a well- controlled manner has attracted a lot of attention in the fields of biosensing and nanodevices. The DNA nanostructures offer addressable substrate for nanoparticle spatial attachment. Our goal is coupling various assembled nanostructures and specific spatial nanoparticle modifications, thus constructing versatile tunable nanoparticle assembly nanostructures.